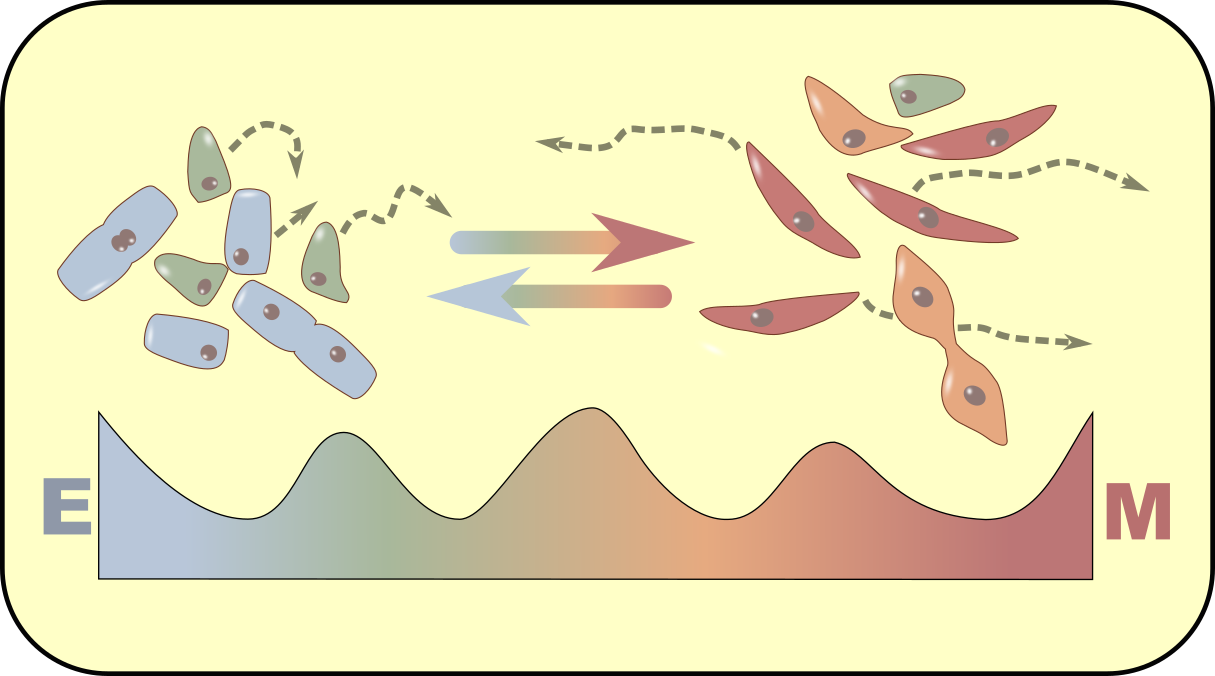
Epithelial-mesenchymal transition
Epithelial-mesenchymal transition is a cellular process that regulate embryonic development, postnatal development, would healing, fibrosis and metastasis. We use mathematical models and statistical approaches to examine the multistate and partially reversible epithelial-mesenchymal transition.
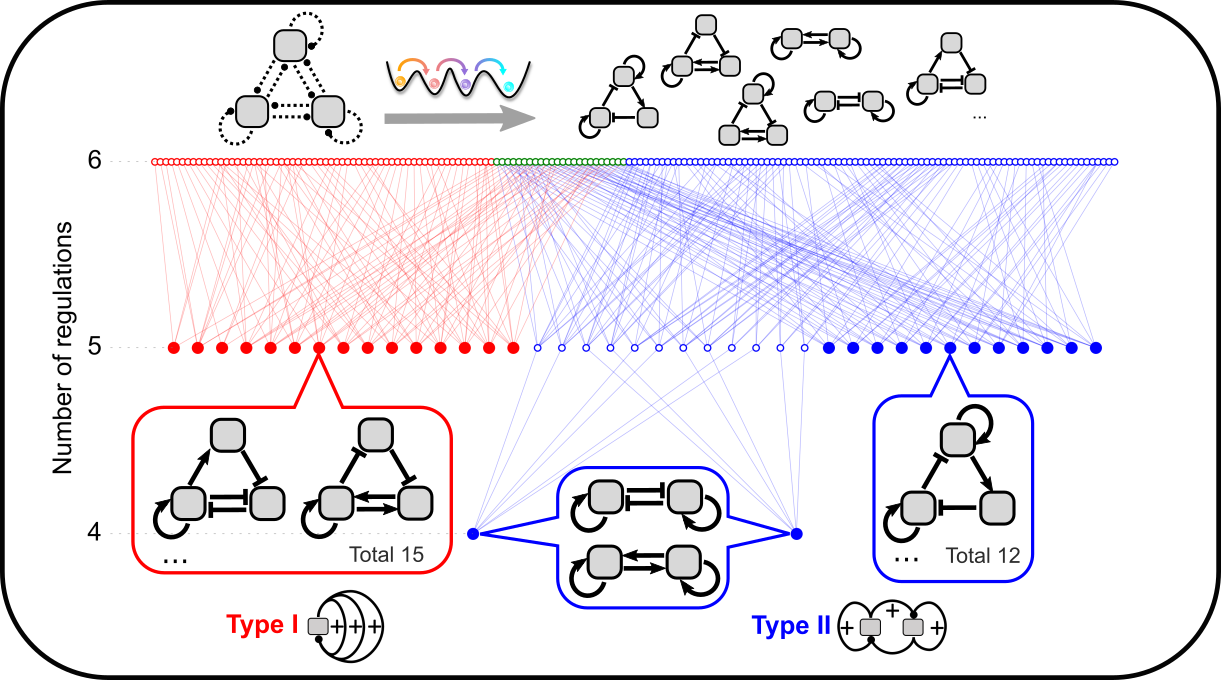
Theories of ultra-high feedbacks and noncanonical feedbacks in gene regulatory networks
Feedback loops in gene regulatory networks have diverse biological functions. We are developing theoretical frameworks to understand the unique functions of highly interconnected feedback loops. We combine mathematical analysis and bioinformatics to connect generic functions of ultra-high feedbacks to real biological networks. We are also interested in systems that have behaviors typically driven by feedback loops but do not appear to have classical feedbacks driven by protein/transcriptional factors.
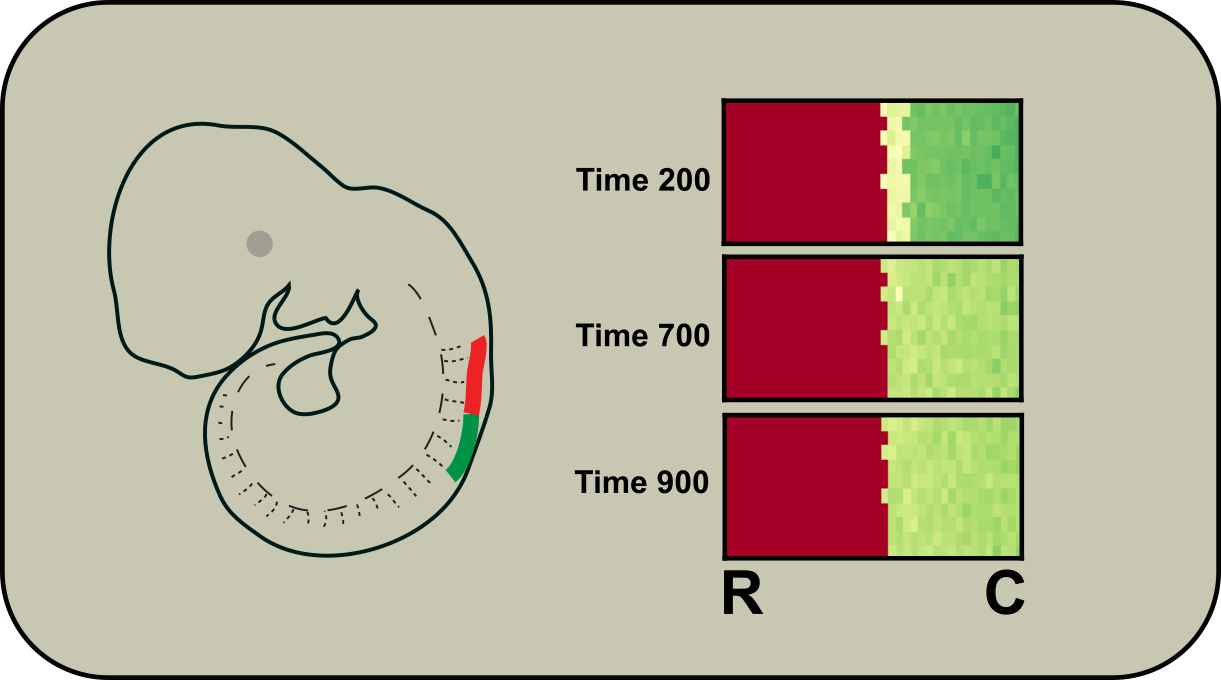
Boundary formation in developing motor neurons
We are interested in pattern formation in development. We are currently focusing on understanding how two types of motor neurons form a sharp boundary during embryogenesis. The boundary underlies highly organized patterns of motor neurons, which play essential roles in specifying their connectivity to diverse muscles accurately. We build models to understand how motor neurons interpret the positional signals with their gene regulatory networks and make fate decisions.
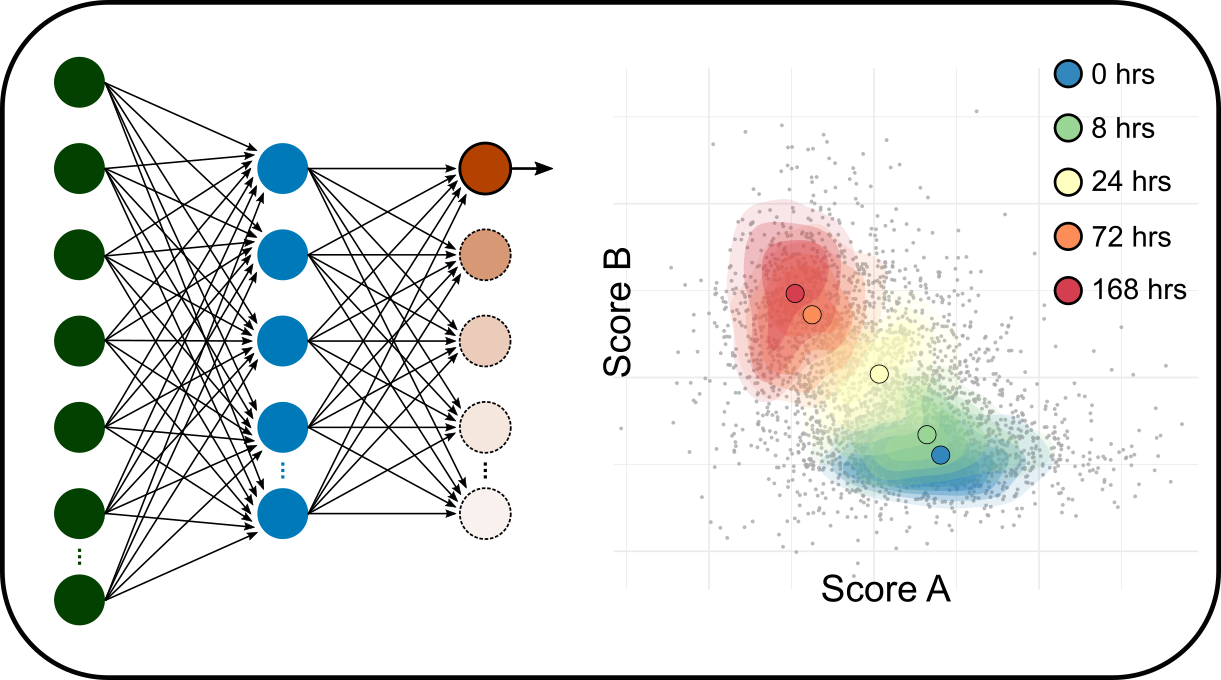
Multi-omics data integration
We are developing computational methods to integrate gene expression (single cell, spatial transcriptome etc.), epigenetics, proteomics and clinical data to interrogate the intricate relationship between dynamics of gene activities and the physiological outcomes.
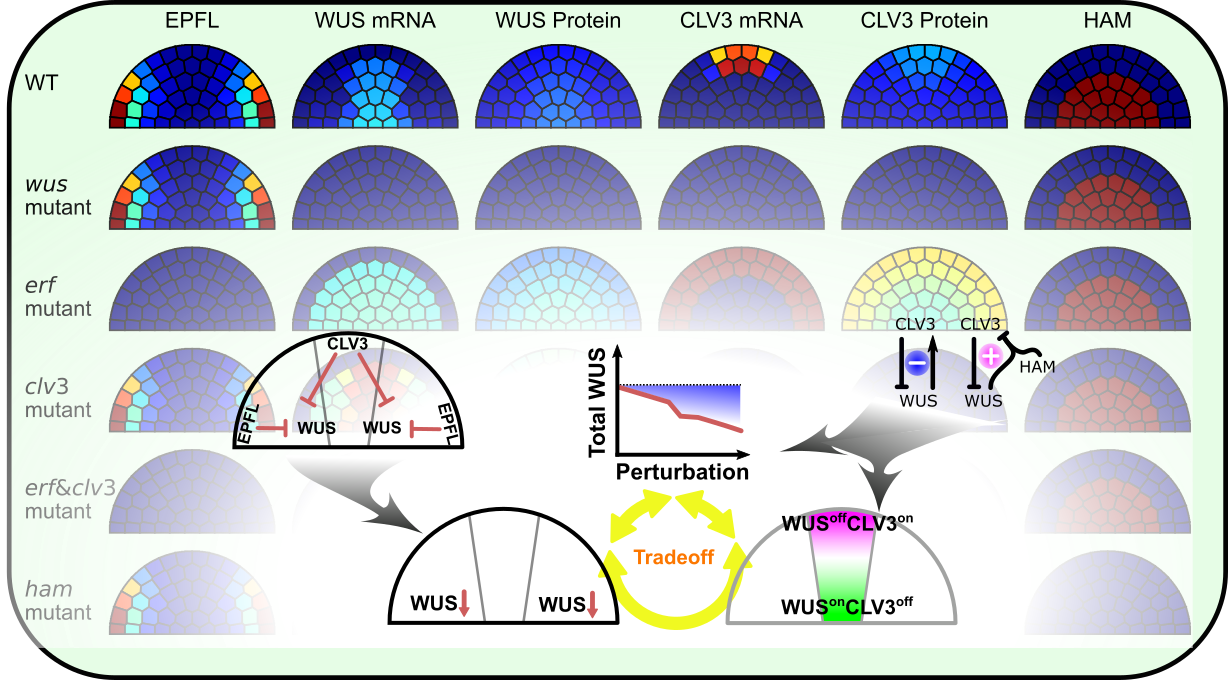
Patterning of the shoot apical meristem
The shoot apical meristem (SAM) is the primary stem cell niche in plant shoots. We are building mathematical models for the regulatory network to understand intriguing and robust pattern formation of the SAM. We are particularly interested in integrations of positional signals at two directions, and paradoxical feedbacks of this system.
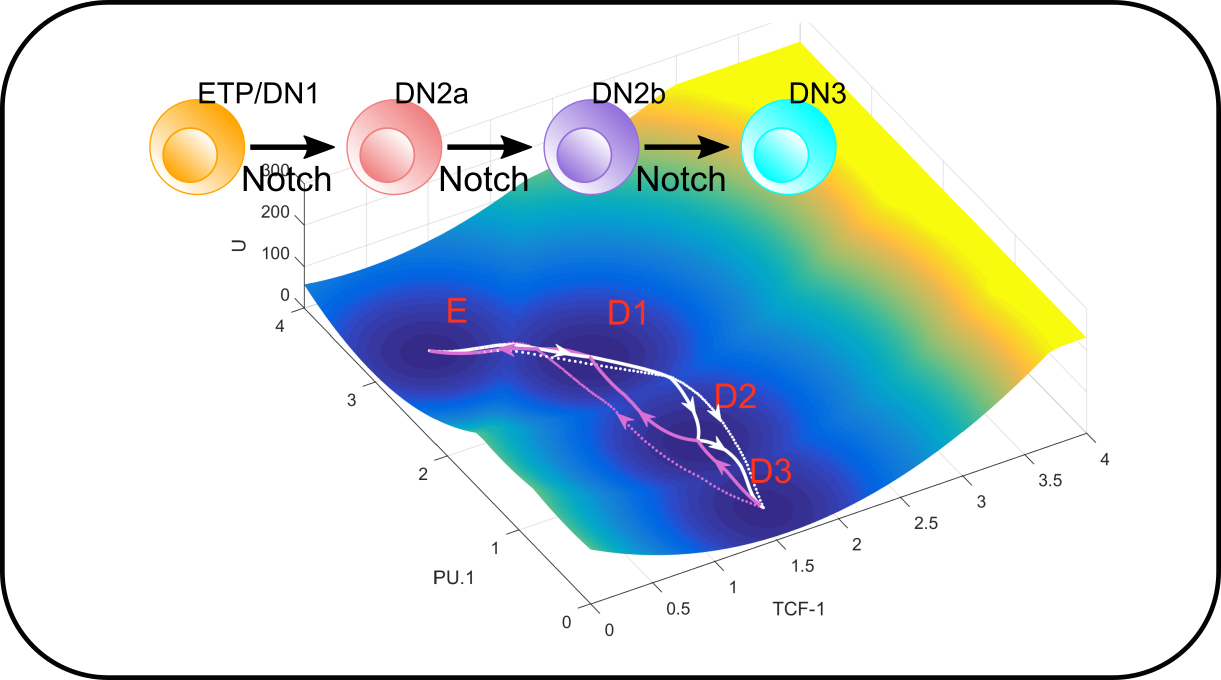
T cell differentiation in development and immune response
We are building models to understand fate transitions in early T cell development as well as terminal lineage specification of functional T cells. In early T cell development, we ask how gene regulatory networks control the progressive and stepwise loss of stemness and commitment to T cell lineage. We are also interested in how T cells diversify themselves during pathogenic challenges, and how the system balances the adaptability of the immune responses and the robustness of lineage specification of individual cells.
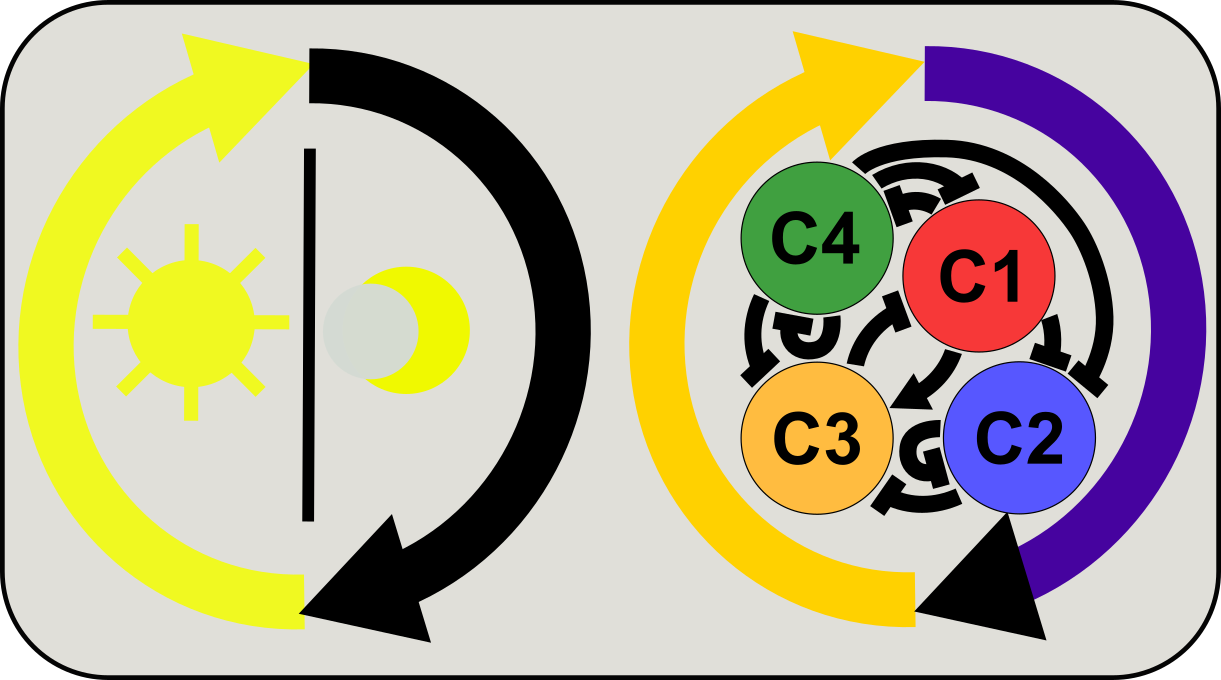
Integration of circadian and diurnal controls of gene/protein activities
The circadian clock provides plants and animals with rhythmic cues to anticipate light-dark (diurnal) cycles. However, diurnal cycles can give rise to cyclic patterns of gene and protein activities in a clock independent manner. We use mathematical modeling and data analysis to understand the signaling and gene expression circuit co-regulated by circadian clock and diurnal cycles, and we are interested in emerging utilities of these intriguing regulatory networks.